OMERO install testing with Docker


The OME team, particularly Jean-Marie and Simon, have been working hard on improving the OMERO install docs.
We have a set of install scripts that include all the commands needed to install and run OMERO, including installation of all dependencies, database set-up and install of OMERO.web with nginx.
These install scripts are used to build our docs for various OSs such as CentOS 7 and Ubuntu.
We can test that the install scripts are working, and then we know that the docs are correct. We can use Docker to do this in a couple of ways:
Automated testing
The omero-install repository has a test script docker-build.sh that creates a Docker image to execute the install scripts in the appropriate OS. There’s also a test_services.sh script that checks OMERO starts up OK and that we can login to OMERO.web.
These tests scripts are run via travis.yml (see builds) so we can see when everything’s green.
But we can also run the docker-build.sh locally, to provide a local OMERO install for testing or evaluation. For example, let’s test OMERO install on Ubuntu 16.04:
$ git clone git@github.com:ome/omero-install.git
$ cd omero-install/linux/test/
$ ./docker-build.sh ubuntu1604_nginx
...
Successfully built 2f3f68971556
Successfully tagged omero_install_test_ubuntu1604_nginx:latest
Test this image by running docker run -it [...] omero_install_test_ubuntu1604_nginxThis installs all the dependencies and creates an omero user who then
installs and runs OMERO.
You should see output of various variables, including the root password:
omero_root_password.
Now we run the Docker container, allowing it to access the host file system and with
various port forwarding in place.
We enter an interactive terminal -it and switch to the omero user who runs OMERO.
Then we can import data from the symlinked directory on my host file system, logging in to OMERO as root user, password omero_root_password:
$ docker run --rm -it -v /Users/wmoore/Documents/biology-data:/biology-data -p 8080:80 -p 4063:4063 -p 4064:4064 omero_install_test_ubuntu1604_nginx
root@c48890350831:/# su omero
omero@c48890350831:/$ cd /home/omero/OMERO.server/
omero@c48890350831:~/OMERO.server$ bin/omero import /biology-data/DV/PTRE/
Server: [localhost:4064]
Username: [omero]root
Password:Then you can open your favorite browser, with your localhost port 8080 forwarding
to port 80 in Docker (as configured above):
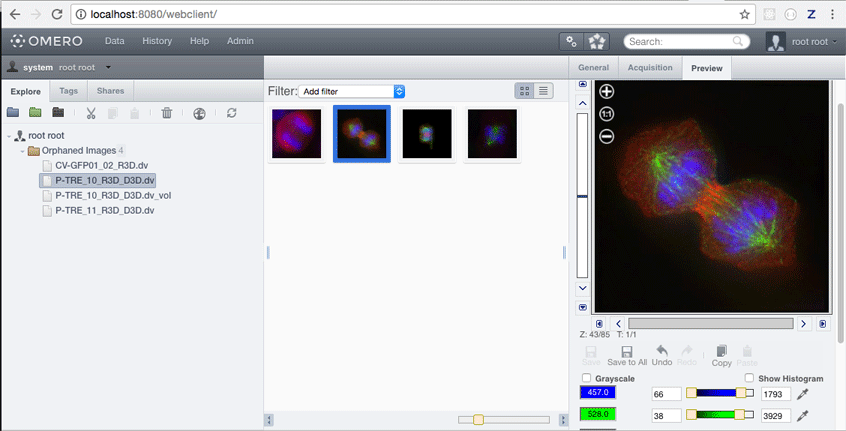
http://localhost:8080/webclient/
Log in as root, omero_root_password and view the imported images.
Manual testing
To test that the auto-generated install docs make sense and provide all the necessary steps, it’s good to follow them manually.
We can use Docker again, this time to provide a clean OS as a starting point. This time we’ll use CentOS 7. We need to allow nginx to access the nextwork, so we use systemd-enabled CentOS 7 with the iproute package, as provided at https://github.com/openmicroscopy/centos-systemd-ip-docker.
$ git clone git@github.com:openmicroscopy/centos-systemd-ip-docker.git
$ cd centos-systemd-ip-docker/
$ docker build ./
...
Successfully built a5f4ebcb0b72This gives us a clean CentOS 7 machine. Now run this and enter the interactive bash terminal.
$ docker run --name c7-systemd -it -d --privileged -p 8080:80 -p 4063:4063 -p 4064:4064 -e LANG=en_US.UTF-8 a5f4ebcb0b72
$ docker exec -ti c7-systemd bin/bashNow you can follow the docs to install OMERO.server on CentOS 7
or install OMERO.web on CentOS 7.
Just remember to run each section as the correct user, $ su omero for the omero user.
Happy testing!